Abstract
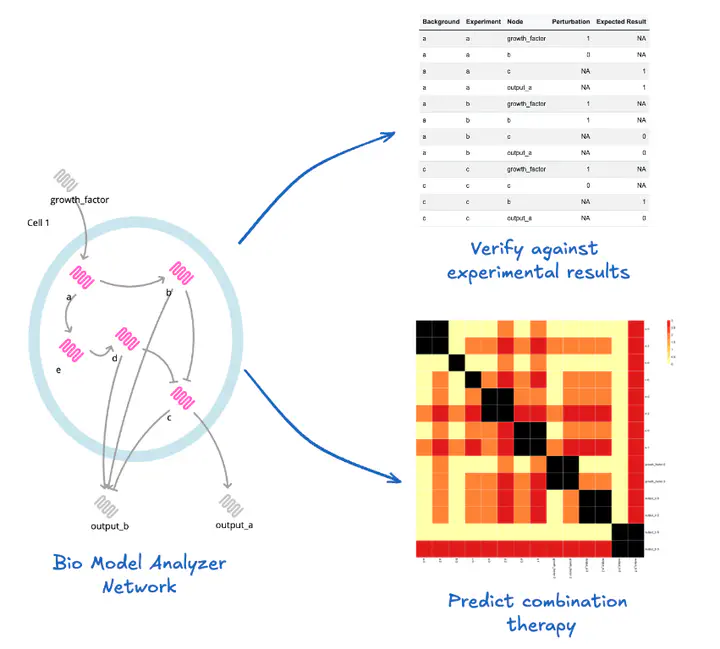
NANSEN (“Network Analysis aNd ScrEeNing”) is an open–source R package that wraps the Bio Model Analyzer (BMA) command-line tools in a tidy, reproducible workflow for verifying, perturbing and systematically screening discrete gene-regulatory network models. The package allows modellers and experimentalists to:
- Validate network specifications – automatically check that experimental perturbation spreadsheets are consistent with the logical model and flag common errors before running long simulations.
- Run large perturbation screens – generate, execute and parse thousands of BMA jobs that test every gene perturbation (“autopert”) or pair-wise gene/drug combination (“combo”).
- Perform high–throughput drug & mutation searches – quantify phenotypic effects of single or double perturbations, optionally against user-defined genetic backgrounds.
- Visualise results – produce publication-ready heat-maps that highlight the most influential nodes, druggable targets or synthetic-lethal pairs.
Type
Publication
Github
Image credit: Adapted from Nansen, 2025