Predicting Cancer Evolution
 CC-BY 4.0
CC-BY 4.0
Cancers evolve from normal cells, and continue to evolve to evade treatment. Understanding evolution from tumourigenesis will allow for earlier detection of cancer, while predicting evolution of resistance mechanisms is essential for finding treatments that are effective in the long term.
We have developed a computational method for efficient search of potential evolutionary pathways from normal cells to cancer and applied this to breast cancer code, paper.
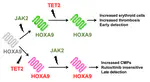
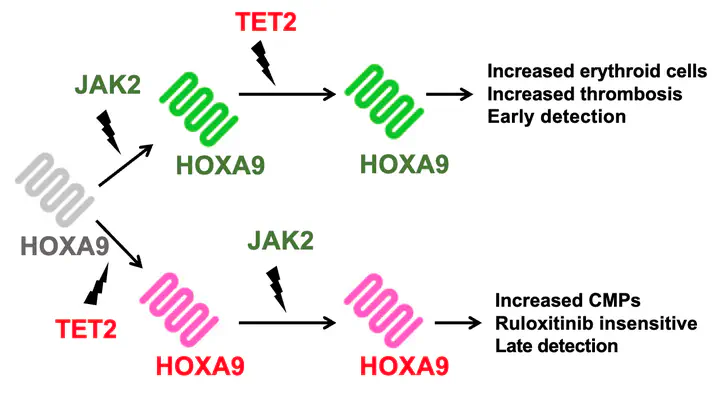
Based on this work, we have also developed a model of myeloproliferative neoplasms (MPNs) that unravels why these neoplasms have different behaviours dependent on the order in which they gain mutations, even if their mutational profiles are otherwise identical paper.
Image credit: Adapted from Talarmain et al., Nature Communications 2022 CC-BY 4.0
Matthew A. Clarke
Research Scientist, Associate Staff Visitor
Research Scientist at the AI Security Institute and Associate Staff Visitor at UCL.