Using State Space Exploration to Determine How Gene Regulatory Networks Constrain Mutation Order in Cancer Evolution
 © Springer Nature Limited 2019
© Springer Nature Limited 2019
Abstract
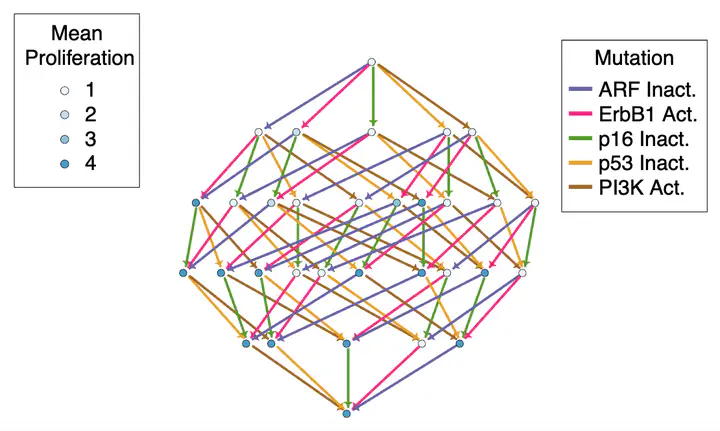
Cancer develops via the progressive accumulation of somatic mutations, which subvert the normal operation of the gene regulatory network of the cell. However, little is known about the order in which mutations are acquired in successful clones. A particular sequence of mutations may confer an early selective advantage to a clone by increasing survival or proliferation, or lead to negative selection by triggering cell death. The space of allowed sequences of mutations is therefore constrained by the gene regulatory network. Here, we introduce a methodology for the systematic exploration of the effect of every possible sequence of oncogenic mutations in a cancer cell modelled as a qualitative network. Our method uses attractor identification using binary decision diagrams and can be applied to both synchronous and asynchronous systems. We demonstrate our method using a recently developed model of ER-negative breast cancer. We show that there are differing levels of constraint in the order of mutations for different combinations of oncogenes, and that the effects of ErbB2/HER2 over-expression depend on the preceding mutations.
Image credit: Adapted from Clarke et al., Automated Reasoning for Systems Biology and Medicine, 2019 (© Springer Nature Limited 2019)